Specifying visual representations for model predictions and observed data using vmc
uncertainty-representation.RmdIntroduction
This vignette describes how to use the vmc package to
specify the visual representations used to describe the distribution of
model predictions and observed data. We developed vmc to
support visual representations of three types: area/extent, visual
variable, and countable icon. For a more general introduction to
vmc and its use on a standard model check workflow, see
vignette("vmc").
Setup
The following libraries are required to run this vignette:
library(dplyr)
library(purrr)
library(vmc)
library(ggplot2)
library(ggdist)
library(cowplot)
library(rstan)
library(brms)
library(gganimate)
library(beeswarm)
theme_set(theme_tidybayes() + panel_border())These options help Stan run faster:
rstan_options(auto_write = TRUE)
options(mc.cores = parallel::detectCores())Model
We use the built-in model mpg_model, a
brmsfit object fitted in Gaussian model family with
push-forward transformations mu and sigma, to
demonstrate different visual representations vmc defines to
show the distribution of model predictions and observed data.
mpg_model
#> Family: gaussian
#> Links: mu = identity; sigma = log
#> Formula: mpg ~ disp + vs + am
#> sigma ~ vs + am
#> Data: mtcars (Number of observations: 32)
#> Draws: 4 chains, each with iter = 6000; warmup = 3000; thin = 1;
#> total post-warmup draws = 12000
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> Intercept 23.25 2.87 17.57 28.92 1.00 6198 6420
#> sigma_Intercept 0.87 0.20 0.50 1.30 1.00 10336 7471
#> disp -0.02 0.01 -0.04 -0.01 1.00 6404 6672
#> vs 2.74 1.74 -0.73 6.14 1.00 6770 6991
#> am 2.74 1.81 -0.76 6.36 1.00 6100 7119
#> sigma_vs 0.27 0.34 -0.38 0.95 1.00 6544 7801
#> sigma_am 0.34 0.36 -0.38 1.03 1.00 6925 7174
#>
#> Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).Visualizing uncertainty using area/extent
vmc supports a list of visual representations to encode
uncertainty using area/extent variable: - mc_model_slab()
(mc_obs_slab()) - mc_model_ccdf()
(mc_obs_ccdf()) - mc_model_cdf()
(mc_obs_cdf()) - mc_model_eye()
(mc_obs_eye()) - mc_model_halfeye()
(mc_obs_halfeye()) - mc_model_histogram()
(mc_obs_histogram()) -
mc_model_pointinterval()
(mc_obs_pointinterval()) - mc_model_interval()
(mc_obs_interval()) - mc_model_lineribbbon()
(mc_obs_lineribbon()) - mc_model_ribbon()
(mc_obs_ribbon())
Examples:
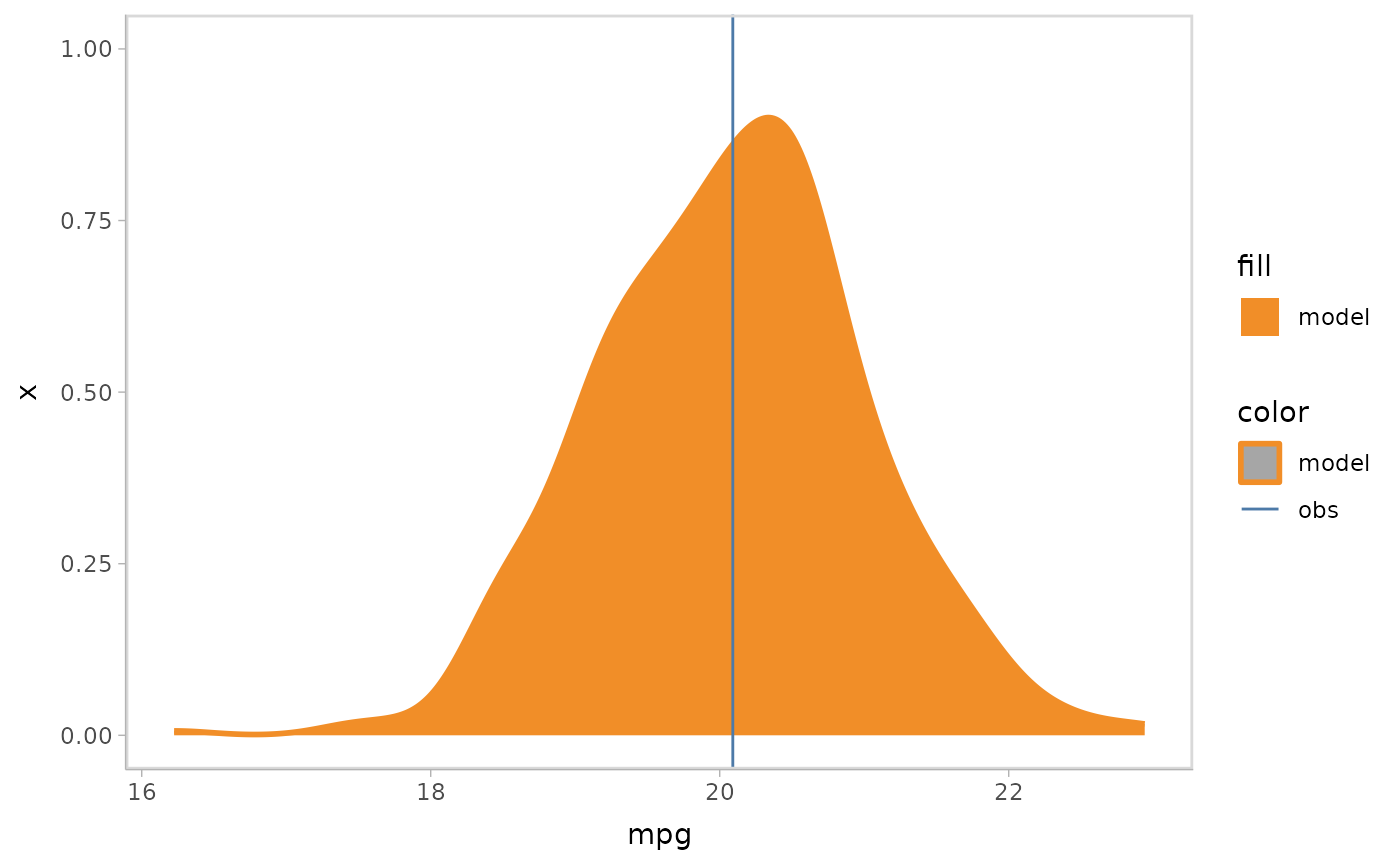
mpg_model %>%
mcplot() +
mc_model_slab(alpha = .5) +
mc_obs_slab(alpha = .5) +
mc_gglayer(coord_flip())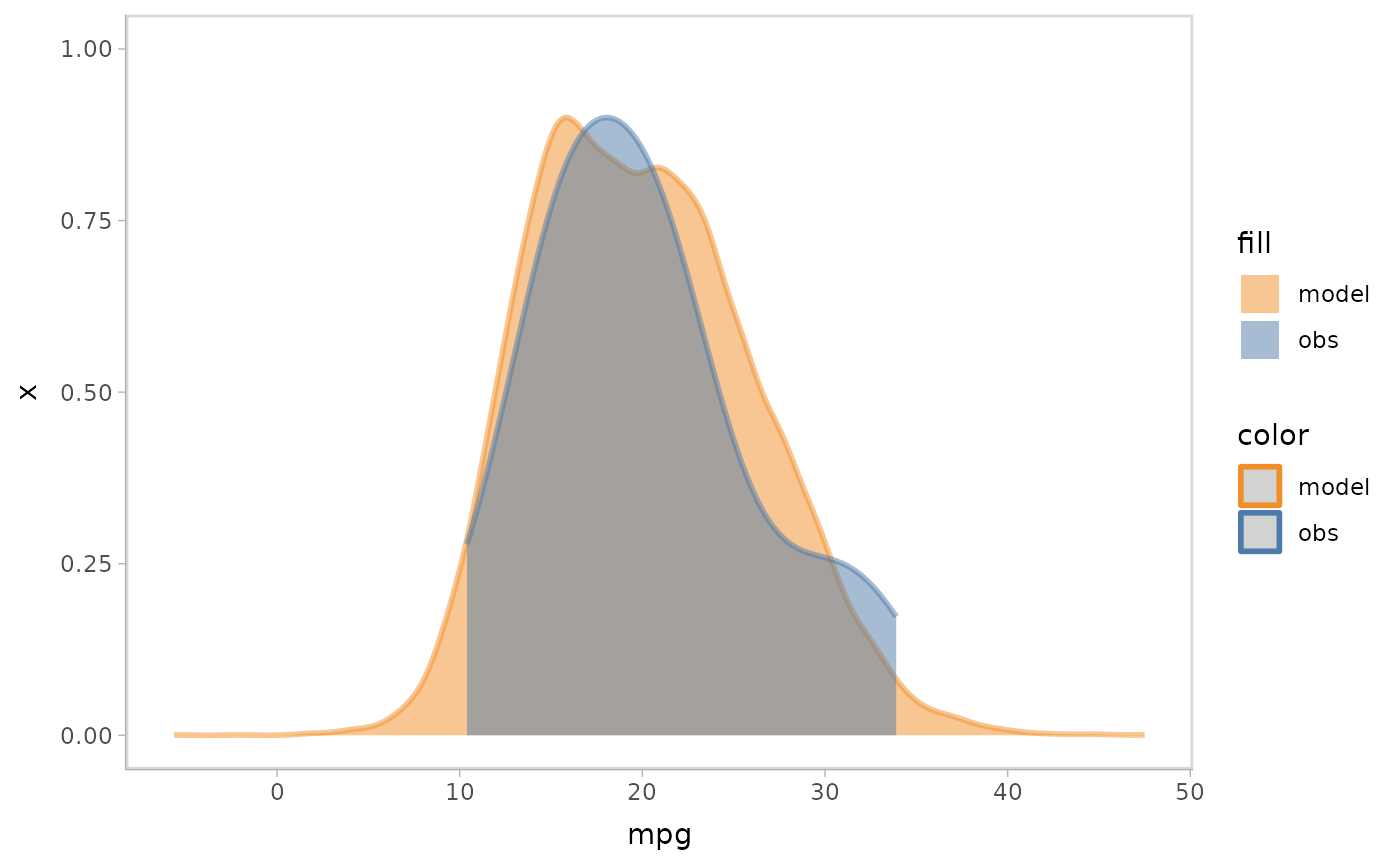
mpg_model %>%
mcplot() +
mc_model_slab(alpha = .5) +
mc_obs_slab(alpha = .5) +
mc_condition_on(x = vars(vs)) +
mc_gglayer(coord_flip())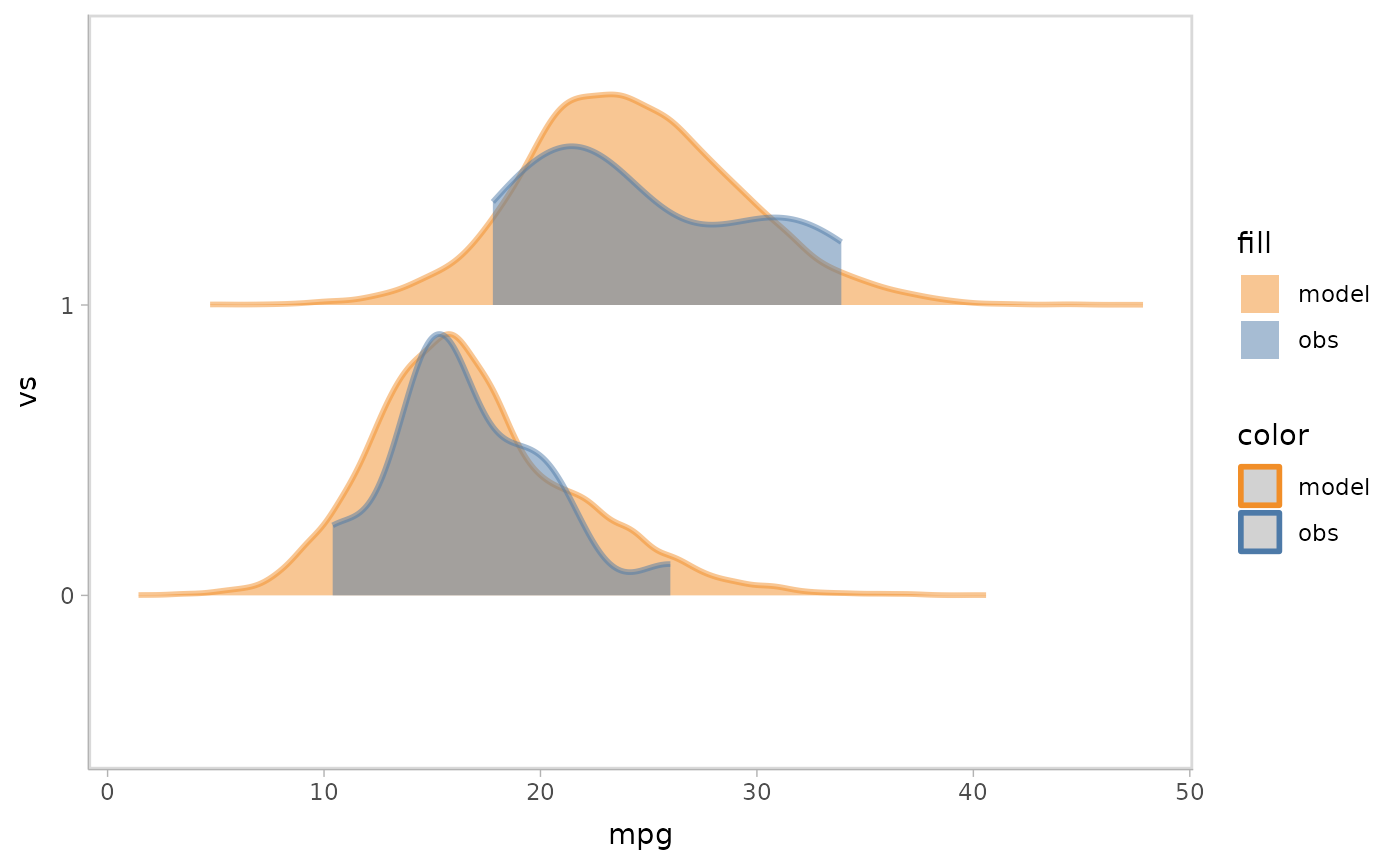
mpg_model %>%
mcplot() +
mc_model_slab(alpha = .5) +
mc_obs_interval(alpha = .5) +
mc_condition_on(x = vars(vs)) +
mc_gglayer(coord_flip())
#> Warning: Duplicated aesthetics after name standardisation: alpha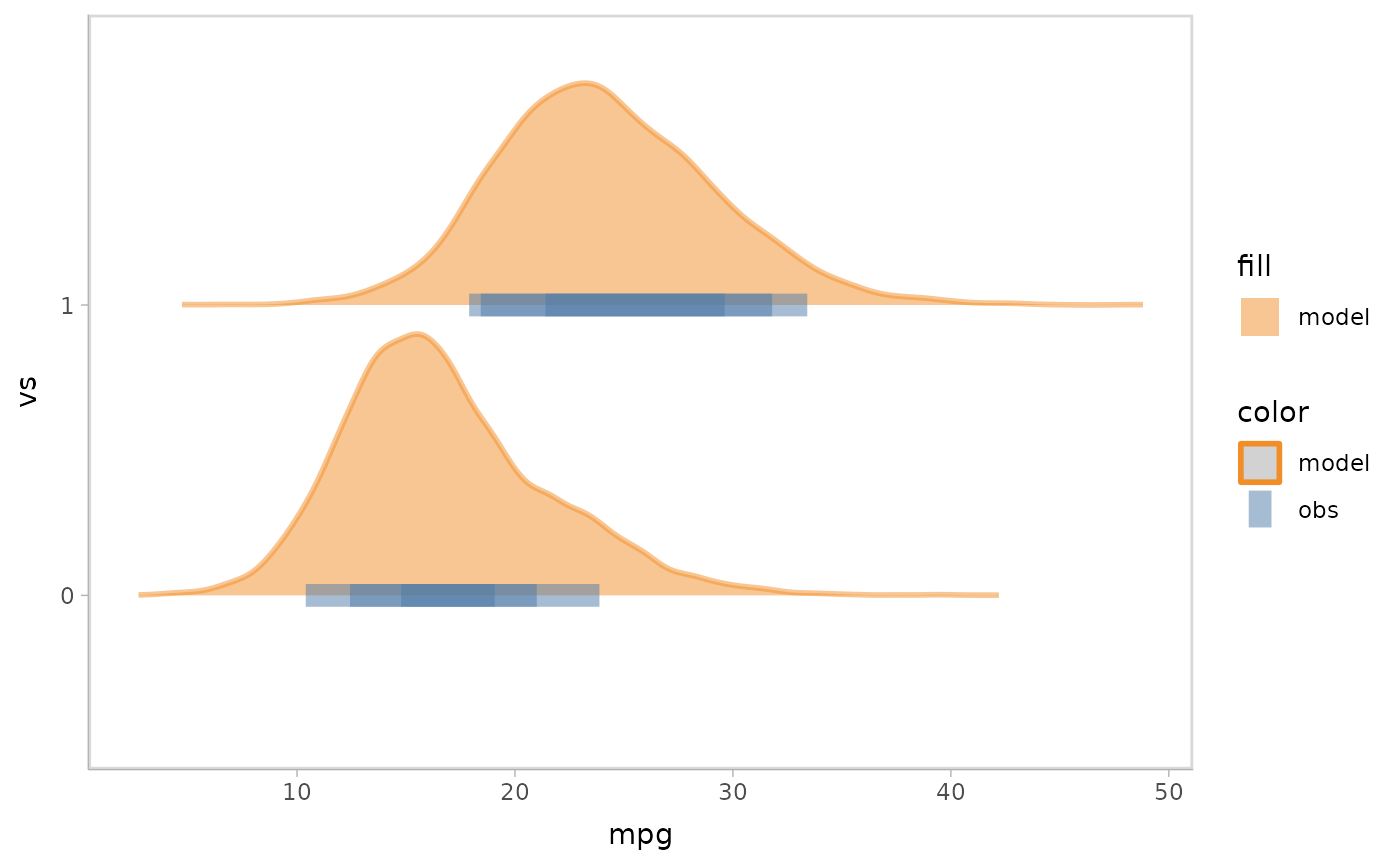
mpg_model %>%
mcplot() +
mc_model_lineribbon() +
mc_obs_pointinterval() +
mc_condition_on(x = vars(disp))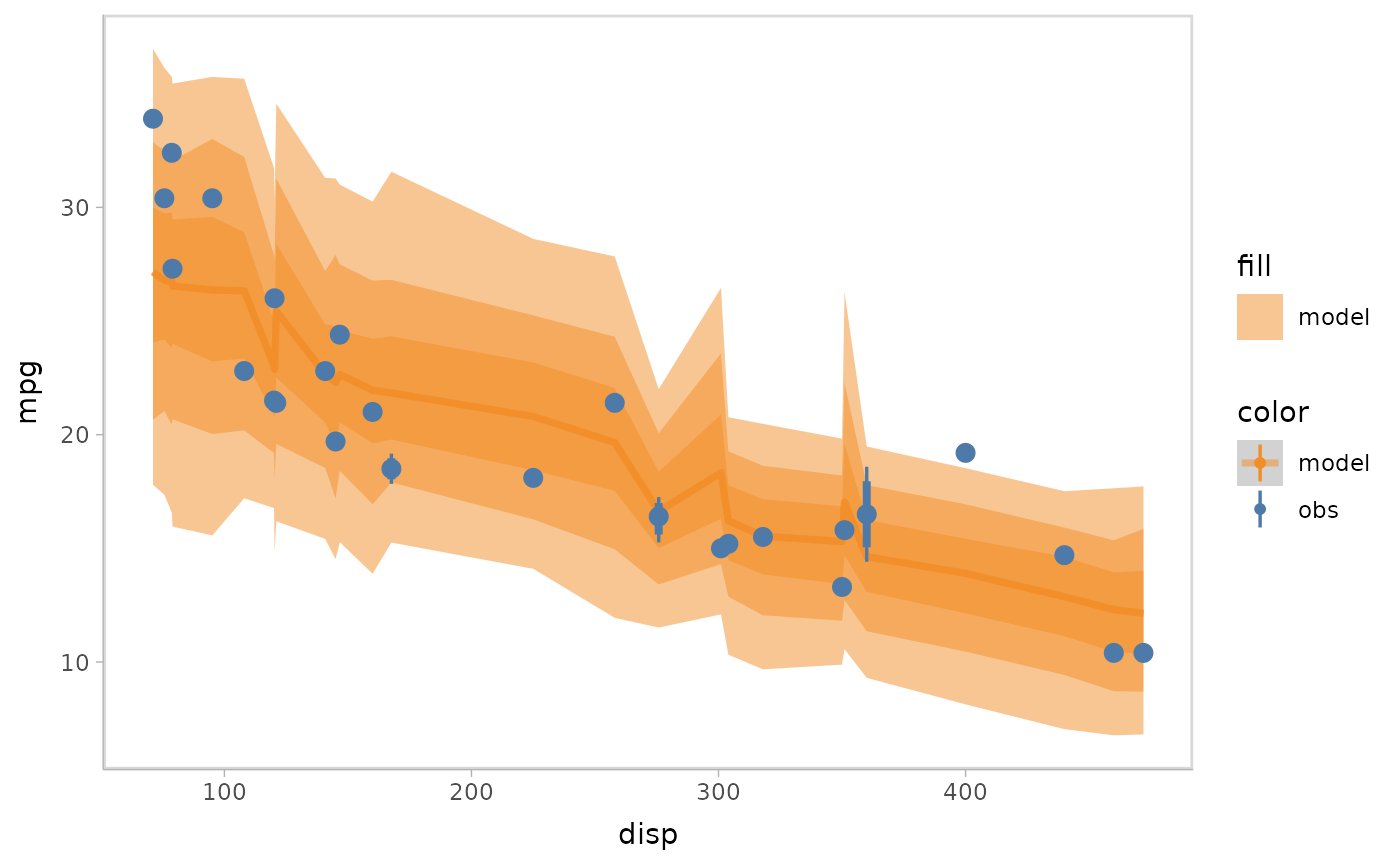
Visualizing uncertainty using visual variables
vmc supports a list of visual representation that encode
uncertainty using diffferent visual variables: -
mc_model_gradientinterval()
(mc_obs_gradientinterval()) -
mc_model_interval() (mc_obs_interval()) -
mc_model_lineribbbon() (mc_obs_lineribbon()) -
mc_model_ribbon() (mc_obs_ribbon()) -
mc_model_tile() (mc_obs_tile())
Examples:
mpg_model %>%
mcplot() +
mc_model_gradientinterval() +
mc_obs_point(shape = '|', size = 10) +
mc_condition_on(x = vars(vs)) +
mc_gglayer(coord_flip())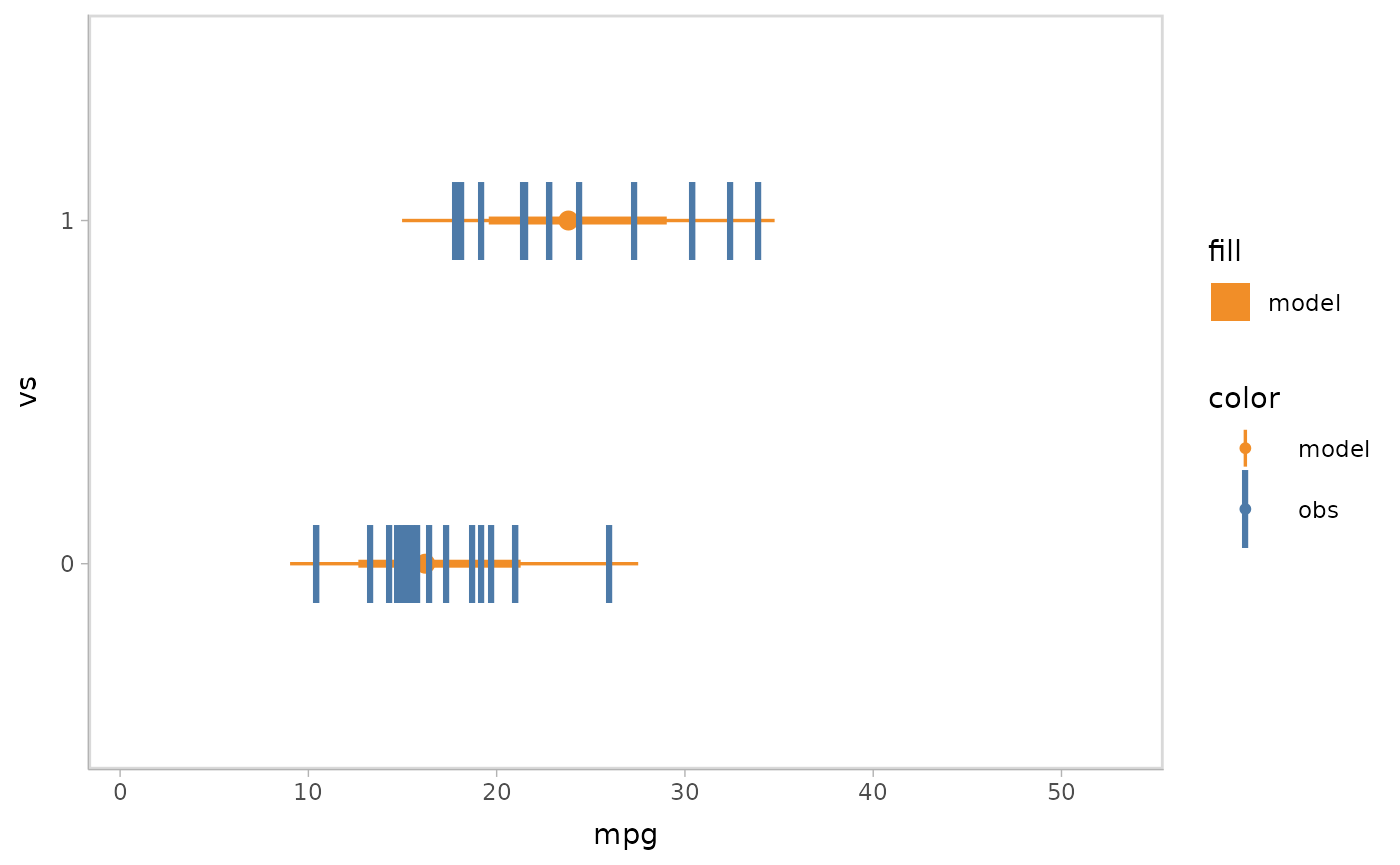
mpg_model %>%
mcplot() +
mc_model_gradientinterval() +
mc_obs_gradientinterval() +
mc_condition_on(x = vars(vs)) +
mc_layout_nested() +
mc_gglayer(coord_flip())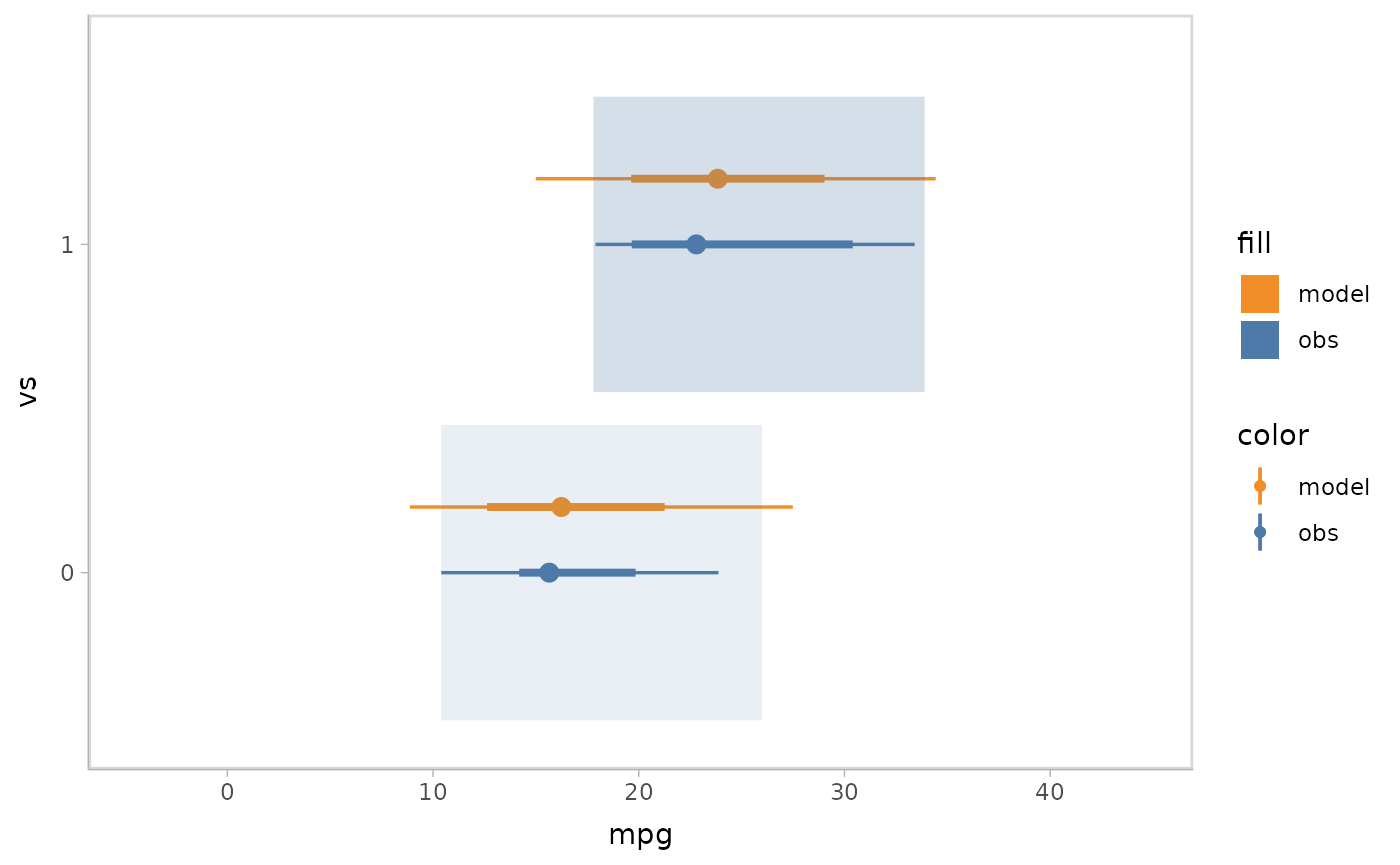
Visualizing uncertainty using countable icons
vmc supports a list of visual representations that
encode uncertainty using countable icons: - mc_model_dots()
(mc_obs_dots()) - mc_model_dotsinterval()
(mc_obs_dotsinterval()) - mc_model_point()
(mc_obs_point()) - mc_model_line()
(mc_obs_line())
Examples:
mpg_model %>%
mcplot() +
mc_model_dots(n_sample = 100) +
mc_obs_dots() +
mc_gglayer(coord_flip())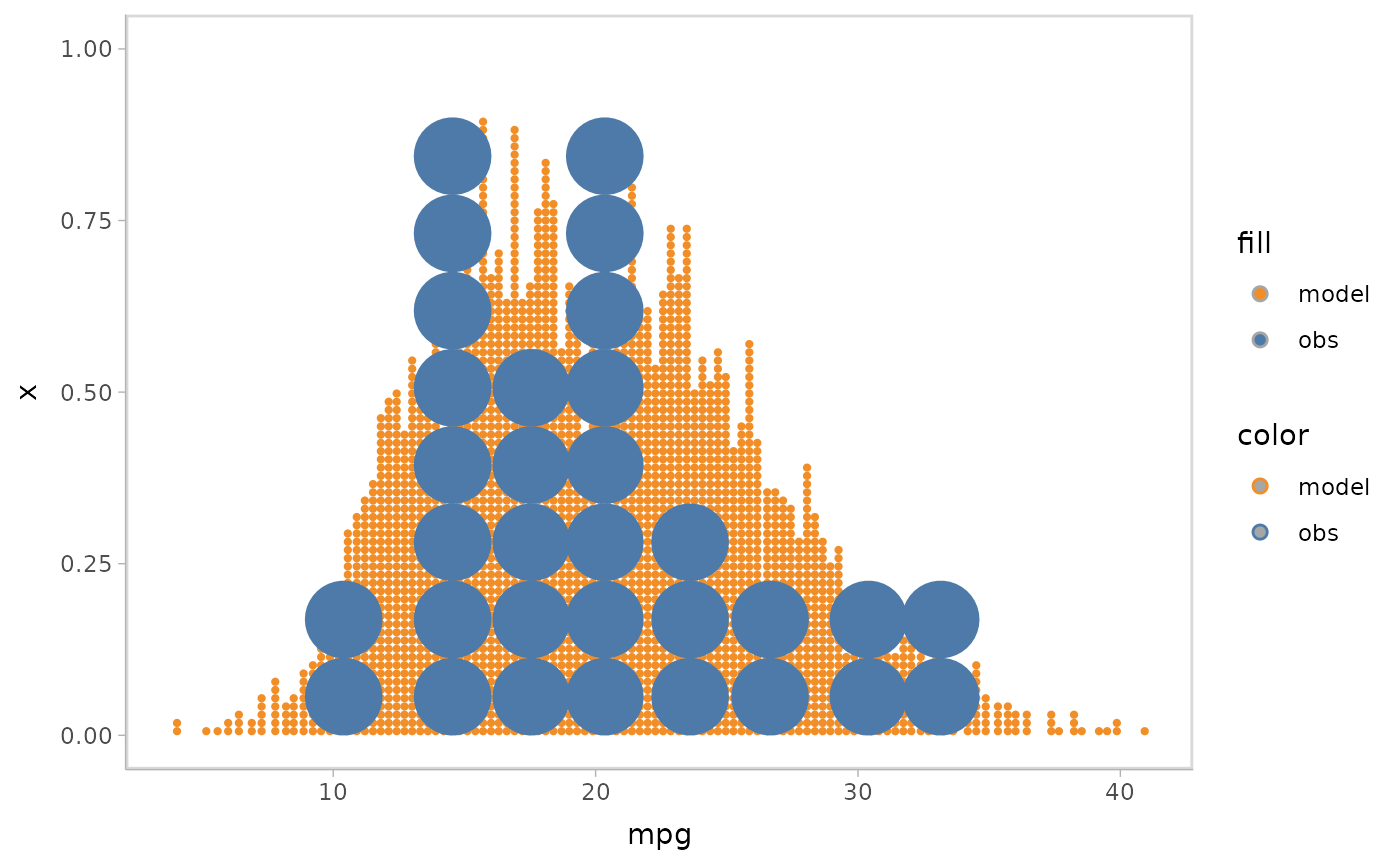
mpg_model %>%
mcplot() +
mc_model_slab() +
mc_obs_interval() +
mc_obs_dots(side = "left") +
mc_condition_on(x = vars(vs)) +
mc_gglayer(coord_flip())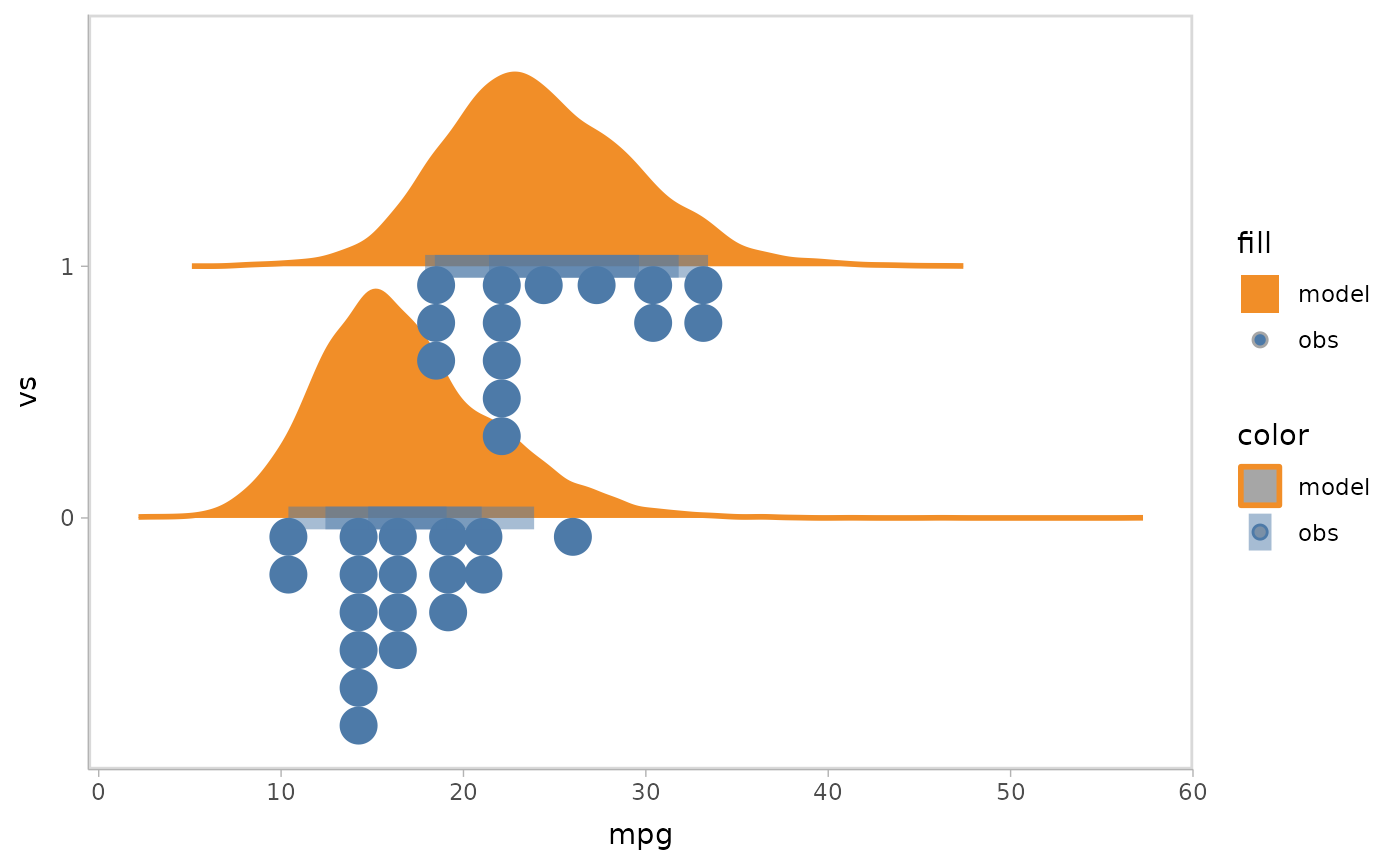
Customize the visual representation
If your visual representation is not supported directly by
vmc, you can use mc_model_custom() and
mc_obs_custom() to pass in a geom or stat function to
specify the visual representation of model predictions and observed
data. For example, we could use
mc_model_custom(stat_boxplot) to specify a box plot for
model predictions and mc_obs_custom(geom_swarm) to specify
a swarm plot for observed data.
mpg_model %>%
mcplot() +
mc_model_custom(stat_boxplot, notch = TRUE) +
mc_obs_custom(geom_swarm) +
mc_condition_on(x = vars(vs)) +
mc_gglayer(coord_flip())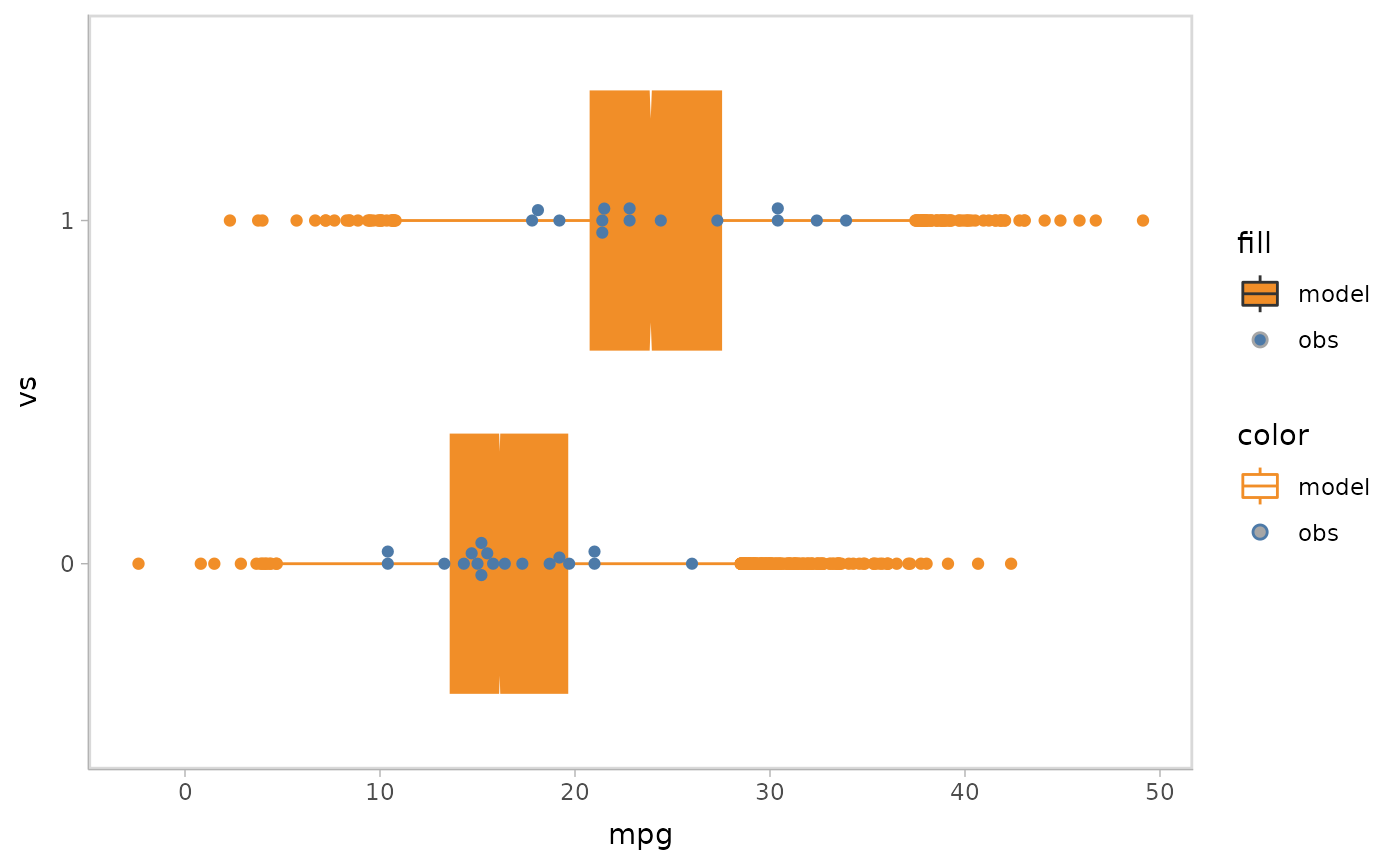
Grouping samples
vmc supports to show the uncertainty information
conditioned on two sources, samples and input data points. Take the slab
representation as example. In the beginning example, we are collapsing
all the samples based on all the input data points together to form one
distribution.
mpg_model %>%
mcplot() +
mc_model_slab(group_sample = "collapse") +
mc_obs_slab() +
mc_gglayer(coord_flip()) +
mc_layout_juxtaposition()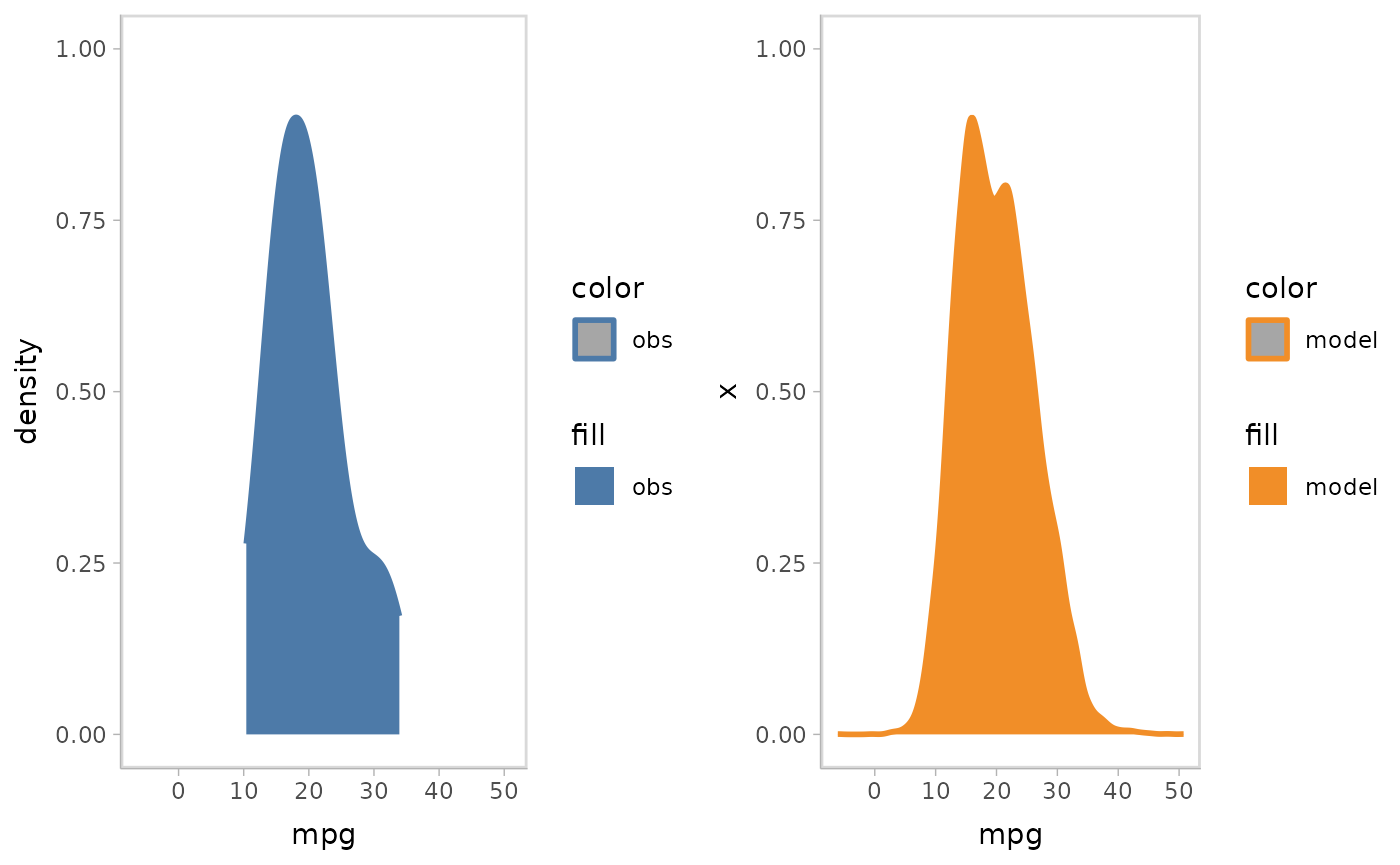
You can also choose to group by the sample when visualizing by slabs (i.e., using individual slabs for each sample).
mpg_model %>%
mcplot() +
mc_model_slab(group_sample = "group", alpha = .2) +
mc_obs_slab() +
mc_gglayer(coord_flip()) +
mc_layout_juxtaposition()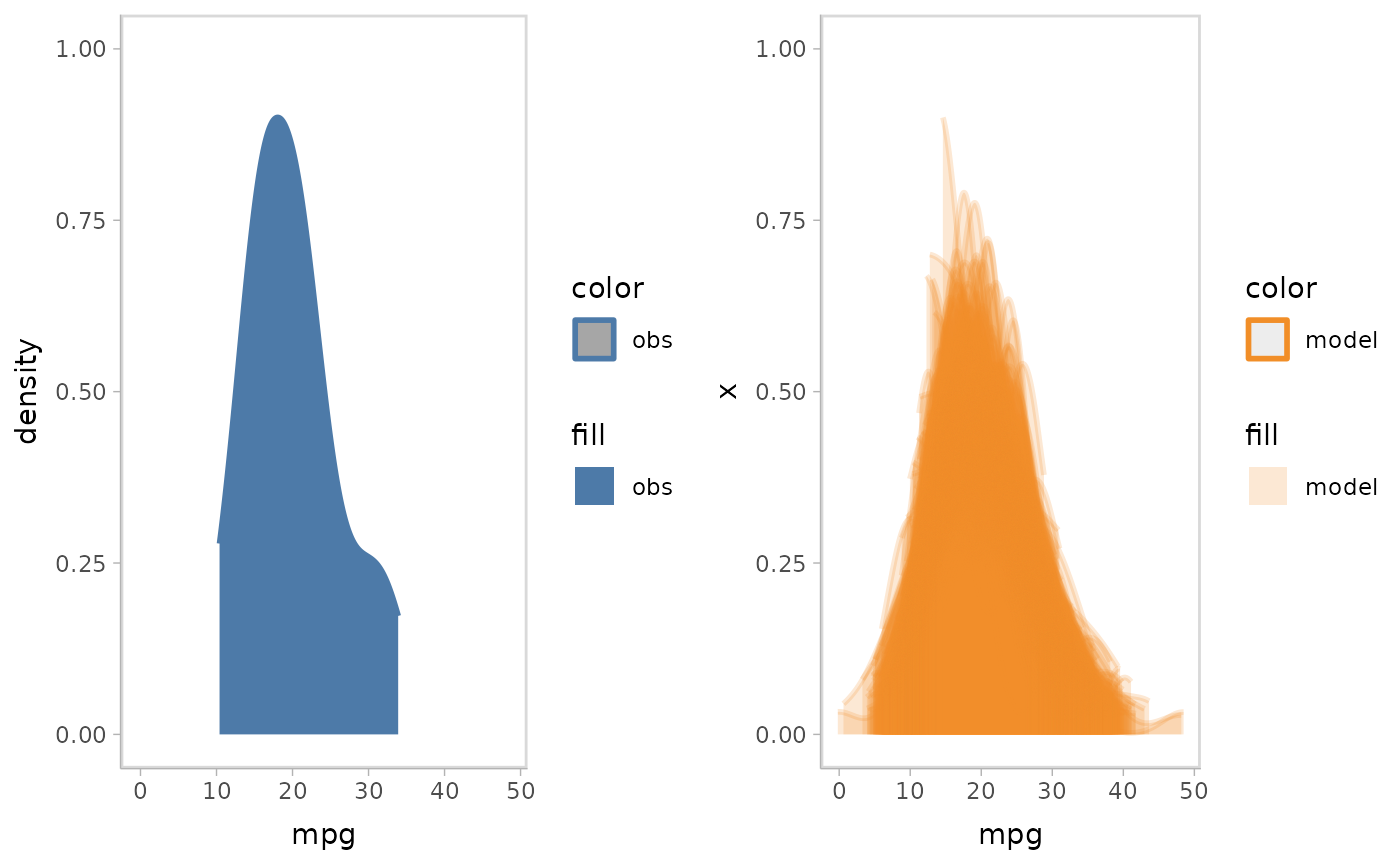
Or you can choose to group on input data point.
mpg_model %>%
mcplot() +
mc_model_slab(group_sample = "group", group_on = "row", alpha = .2) +
mc_obs_slab() +
mc_gglayer(coord_flip()) +
mc_layout_juxtaposition()
#> Warning in layer_slabinterval(data = data, mapping = mapping, stat = StatSlab,
#> : Ignoring unknown parameters: `group_on`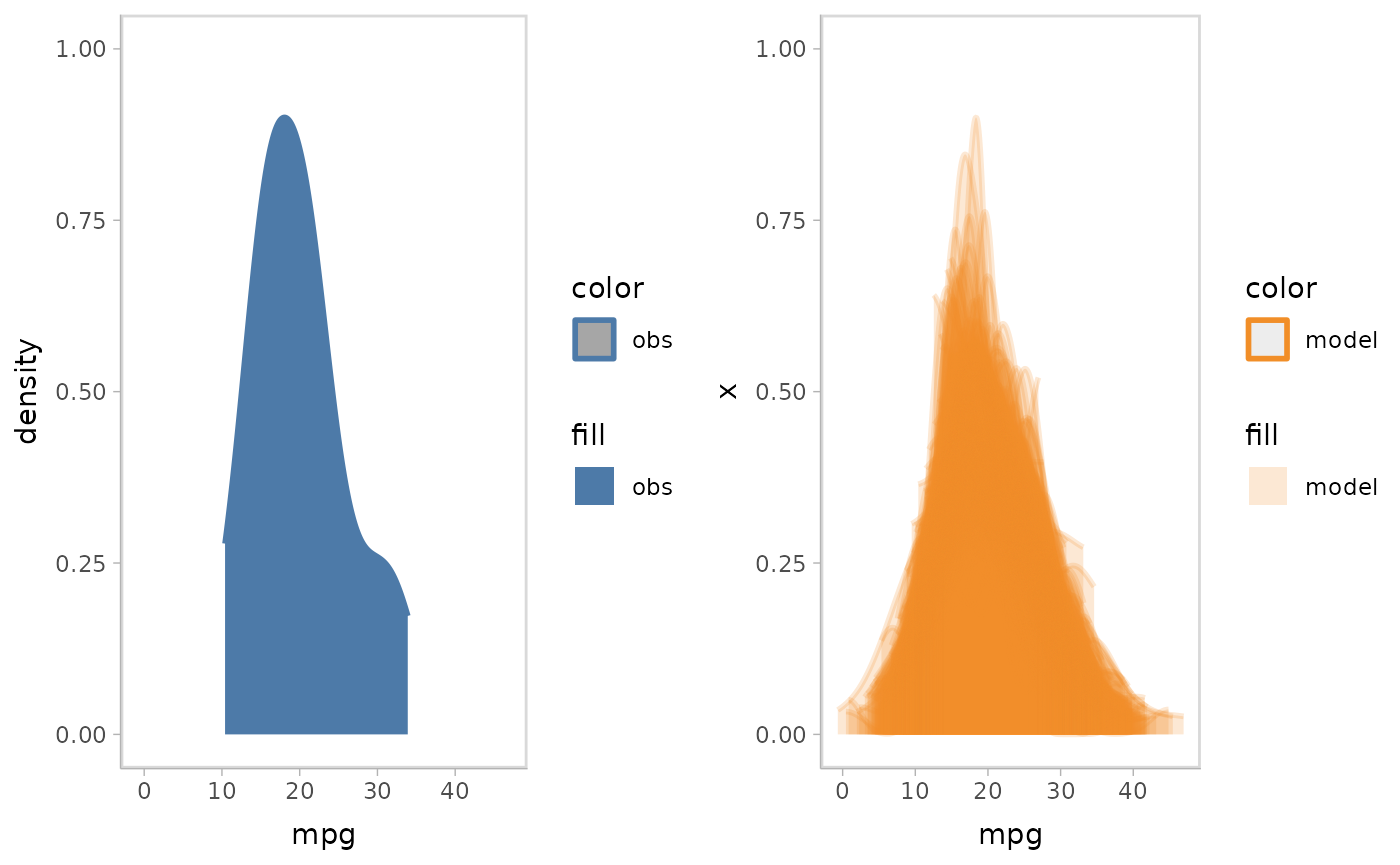
Instead of overlapping all samples or rows together, you can choose to use HOPs to show them each in one time frame.
mpg_model %>%
mcplot() +
mc_draw(ndraws = 50) +
mc_model_slab(group_sample = "hops") +
mc_obs_slab(alpha = .5) +
mc_gglayer(coord_flip())
#> `nframes` and `fps` adjusted to match transition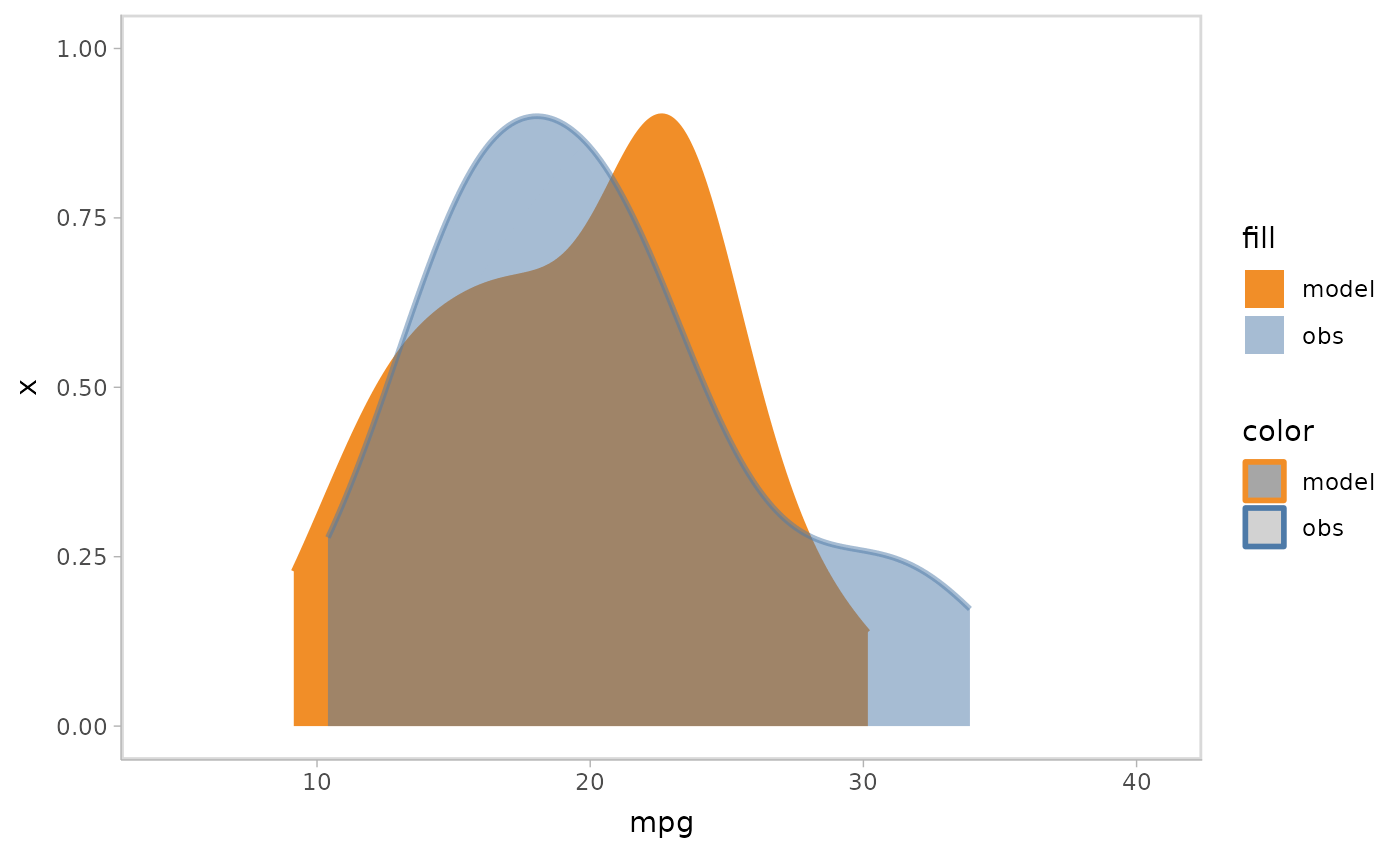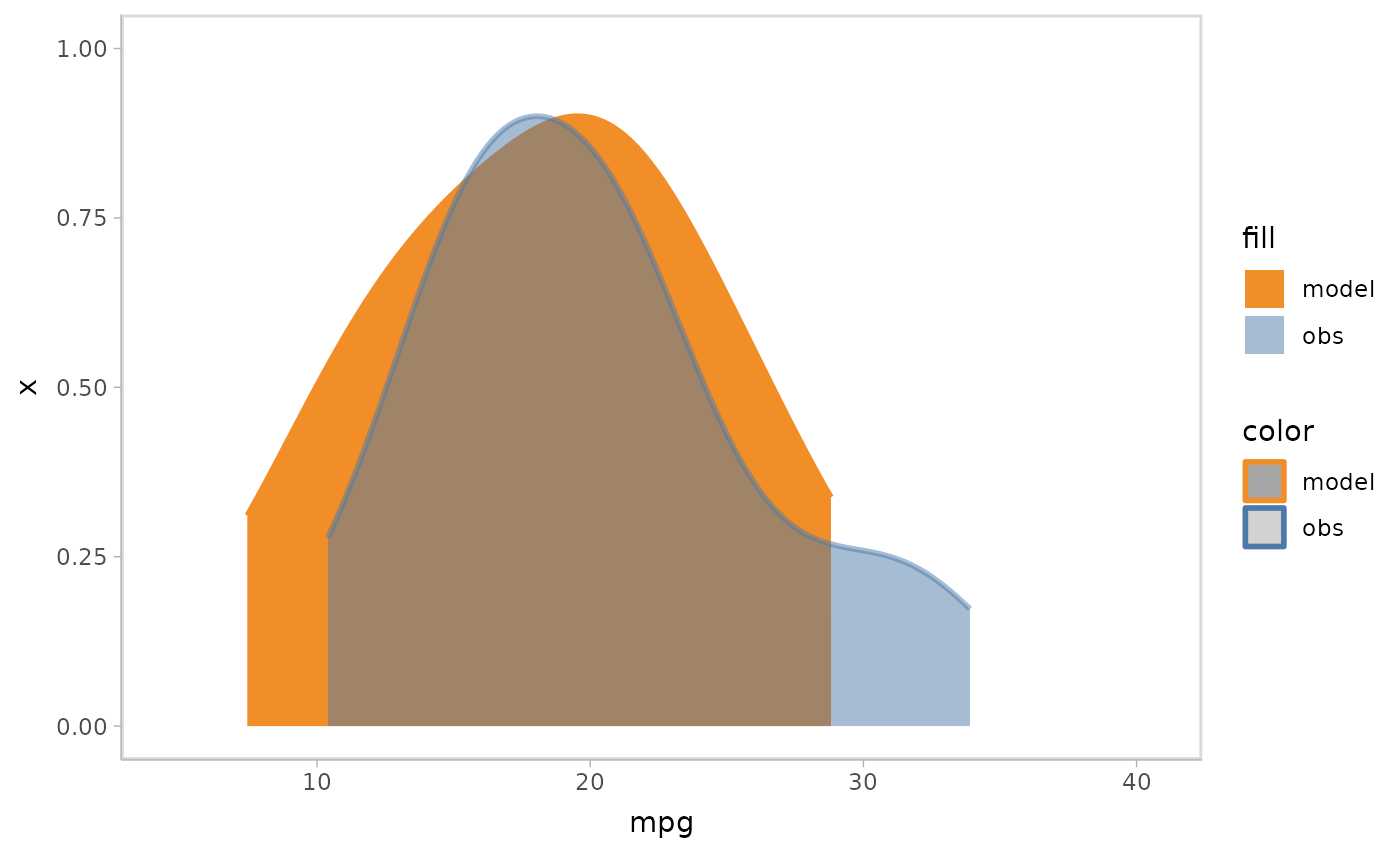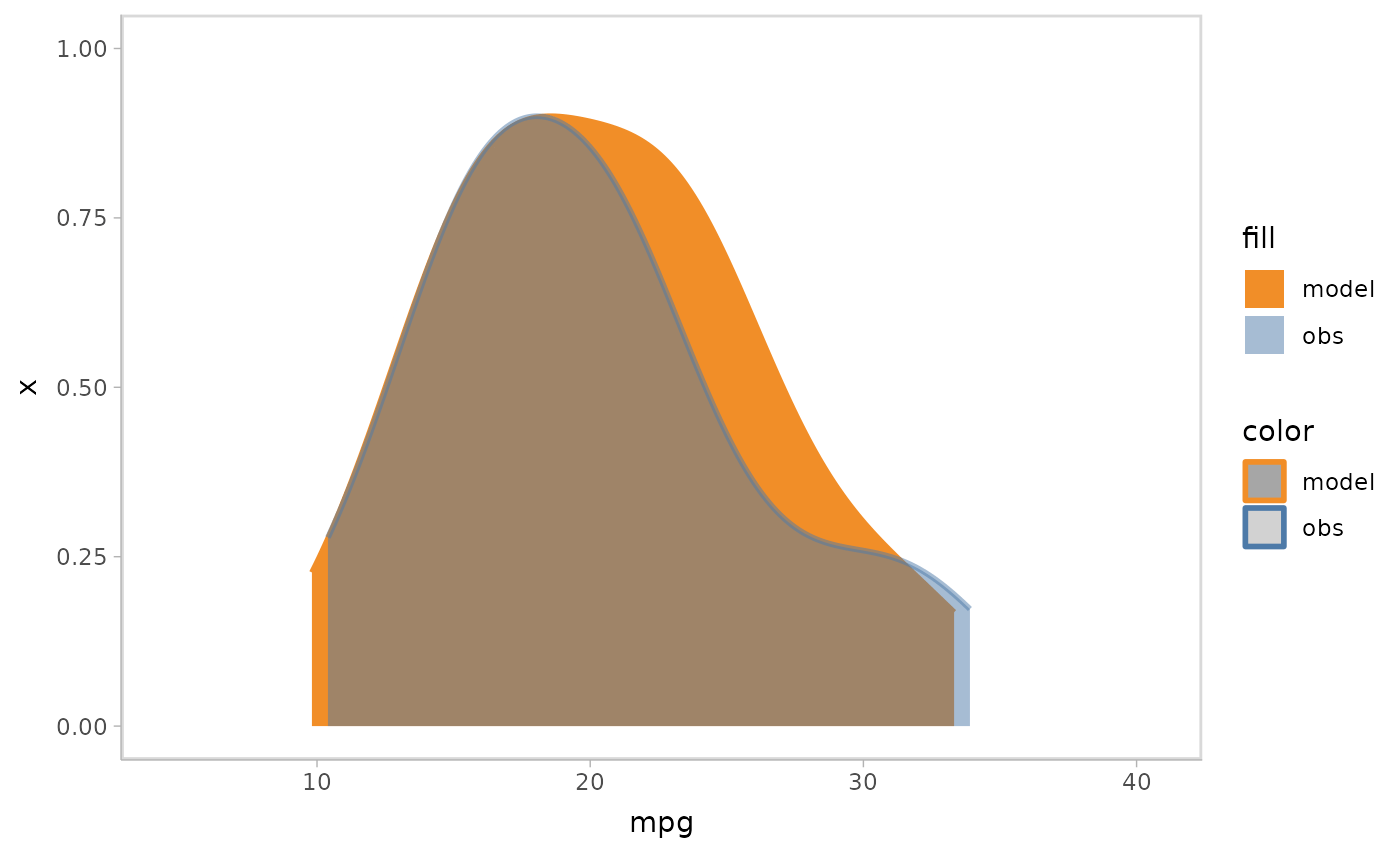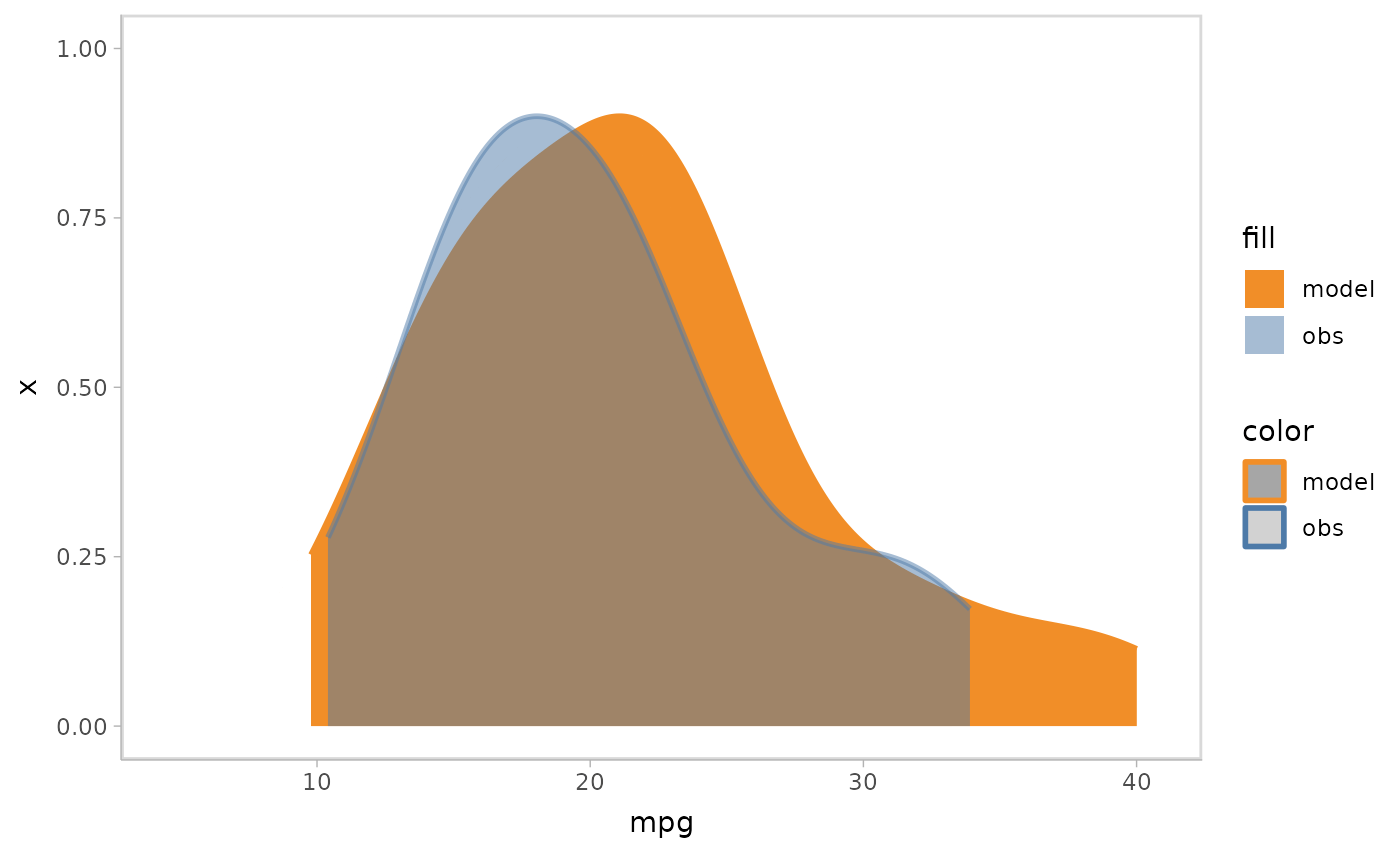
You can also aggregate the model predictions from samples or input data points by functions.
mpg_model %>%
mcplot() +
mc_model_slab(group_sample = mean) +
mc_obs_slab(alpha = .5) +
mc_gglayer(coord_flip())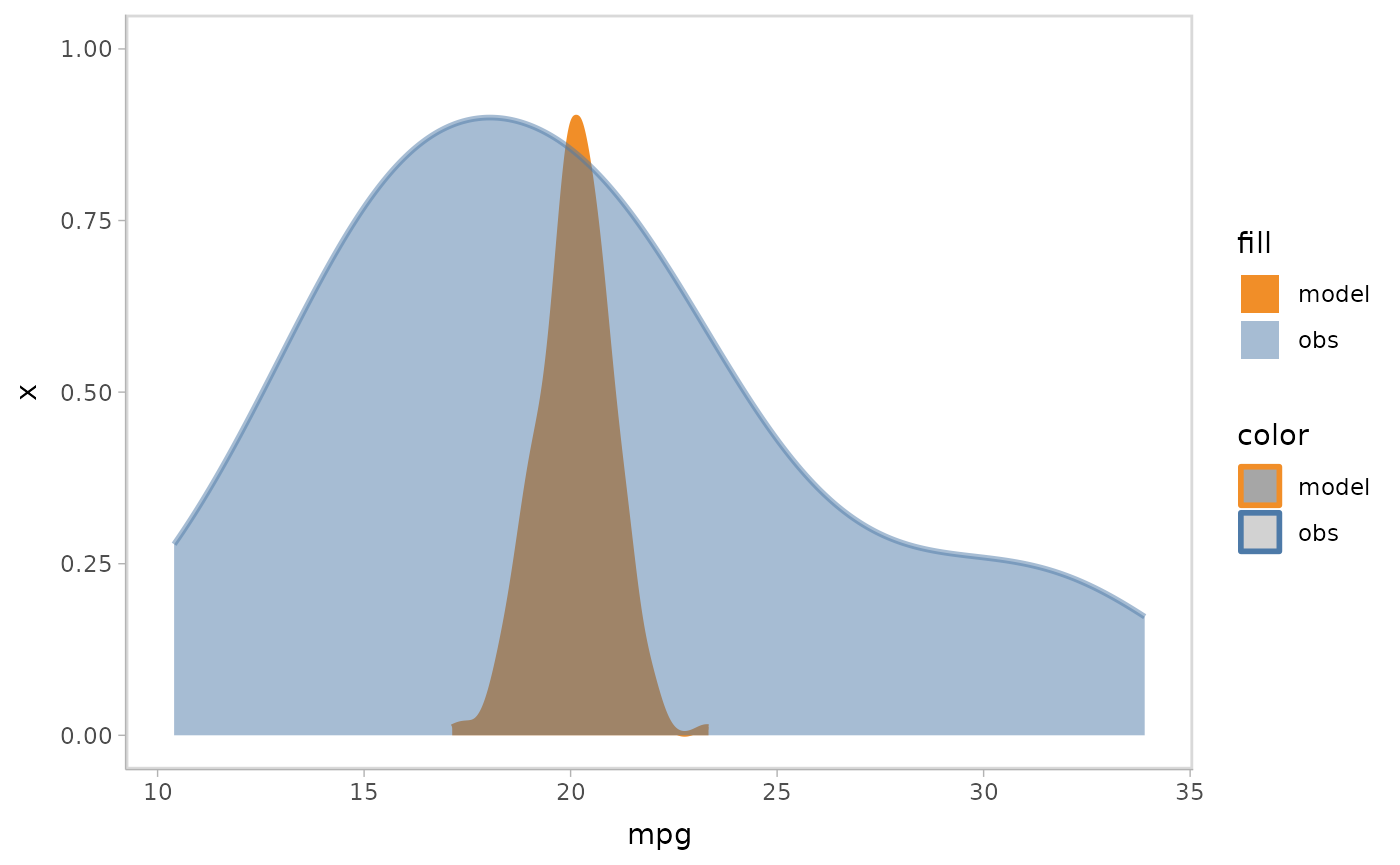
mpg_model %>%
mcplot() +
mc_observation_transformation(mean) +
mc_model_slab(group_sample = mean, group_on = "row") +
mc_obs_reference_line() +
mc_gglayer(coord_flip())
#> Warning in layer_slabinterval(data = data, mapping = mapping, stat = StatSlab,
#> : Ignoring unknown parameters: `group_on`