Comparing model predictions and observed data using vmc
comparative-layout.RmdIntroduction
This vignette describes how to use the vmc package to
specify a comparative layout that organizes the visual representation of
model predictions and observed data into a layout to enhance visual
comparison. We developed vmc to support comparative layouts
of three types: juxtaposition, superposition, and explicit encoding. For
a more general introduction to vmc and demonstration of its
use in a standard model check workflow, see
vignette("vmc").
Setup
The following libraries are required to run this vignette:
library(dplyr)
library(purrr)
library(vmc)
library(ggplot2)
library(ggdist)
library(cowplot)
library(rstan)
library(brms)
library(gganimate)
theme_set(theme_tidybayes() + panel_border())These options help Stan run faster:
rstan_options(auto_write = TRUE)
options(mc.cores = parallel::detectCores())Model
<<<<<<< HEAD We use the built-in model
mpg_model, a brmsfit object fitted in Gaussian
model family with push-forward transformations mu and
sigma, to demonstrate different visual representations
vmc defines to show the distributions in model and observed
data. ======= We use the built-in model mpg_model, a
brmsfit object fitted in Gaussian model family with
checkable push-forward transformations mu and
sigma, to demonstrate different visual representations that
vmc defines to show the distributions of model predictions
and observed data. >>>>>>> main
mpg_model
#> Family: gaussian
#> Links: mu = identity; sigma = log
#> Formula: mpg ~ disp + vs + am
#> sigma ~ vs + am
#> Data: mtcars (Number of observations: 32)
#> Draws: 4 chains, each with iter = 6000; warmup = 3000; thin = 1;
#> total post-warmup draws = 12000
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> Intercept 23.25 2.87 17.57 28.92 1.00 6198 6420
#> sigma_Intercept 0.87 0.20 0.50 1.30 1.00 10336 7471
#> disp -0.02 0.01 -0.04 -0.01 1.00 6404 6672
#> vs 2.74 1.74 -0.73 6.14 1.00 6770 6991
#> am 2.74 1.81 -0.76 6.36 1.00 6100 7119
#> sigma_vs 0.27 0.34 -0.38 0.95 1.00 6544 7801
#> sigma_am 0.34 0.36 -0.38 1.03 1.00 6925 7174
#>
#> Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).Juxtaposition
Juxtaposition puts the visual representations of model predictions
and observed data side by side. This comparative layout presents each
object (visual representation for model predictions and observed data)
separately, but requires that the viewer scan the plots to relate them.
We specify the juxtaposition layout for a model check by
mc_layout_juxtaposition().
mpg_model %>%
mcplot() +
mc_layout_juxtaposition() +
mc_gglayer(coord_flip())
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.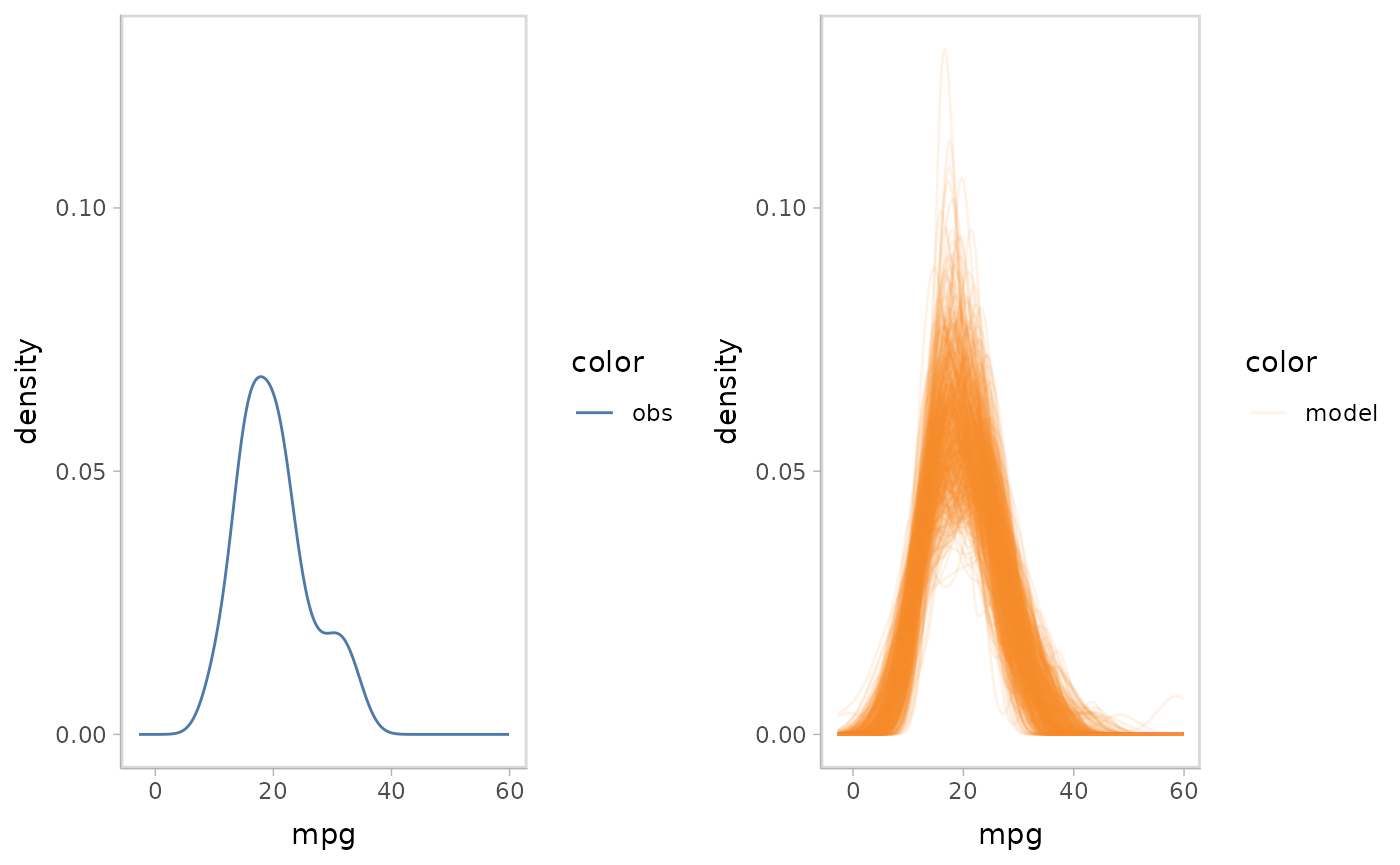
vmc also supports a variant of juxtaposition, nested
juxtaposition, which helps reduce the amount of scanning required by
viewers by nesting the juxtaposition layout in the plot when the model
check has a discrete conditional variable. We specify this by
mc_layout_nested().
mpg_model %>%
mcplot() +
mc_condition_on(x = vars(vs)) +
mc_model_auto(n_sample = 1) +
mc_layout_nested()
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.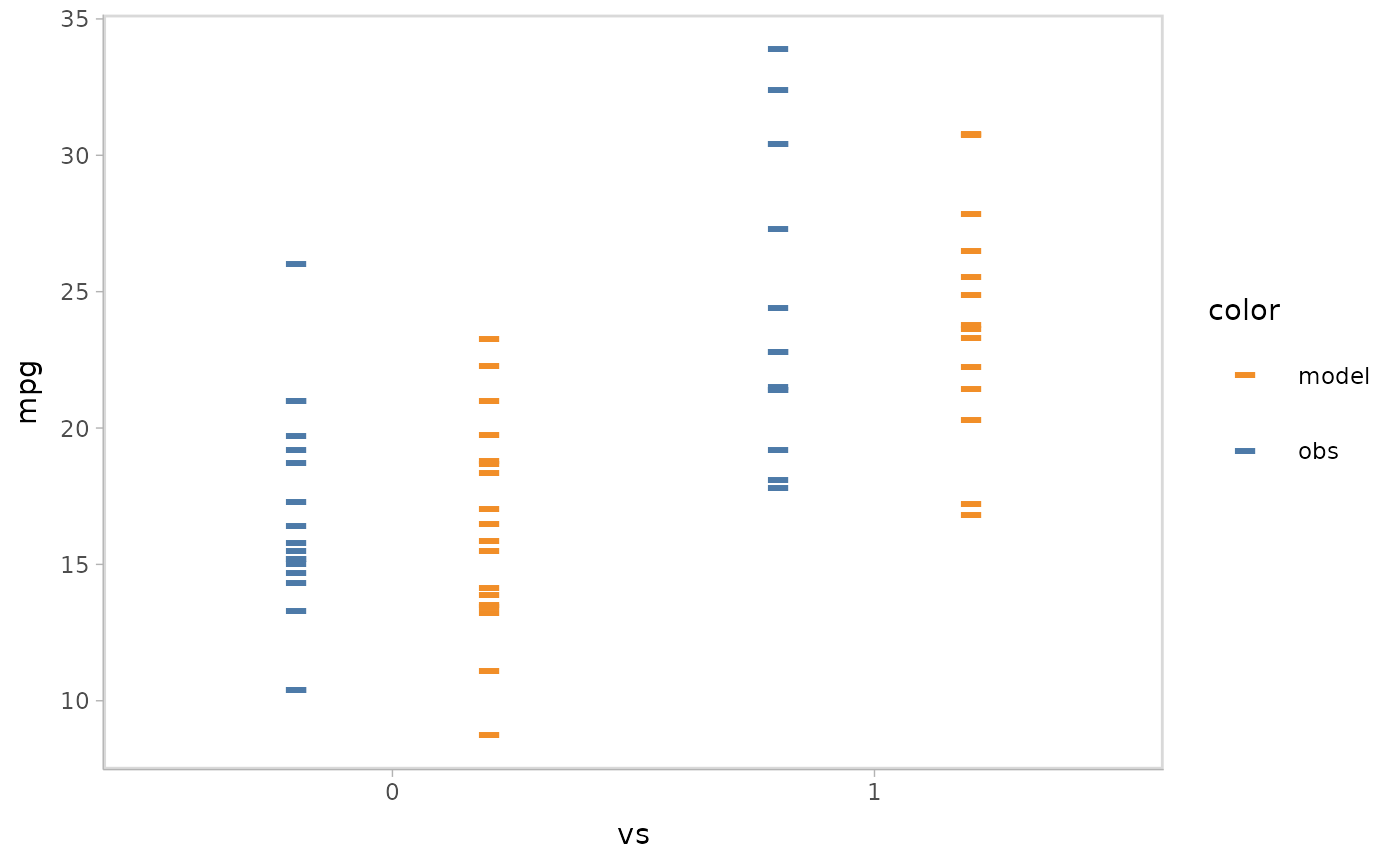
Superposition
Superposition overlays the objects (visual representations for model
predictions and observed data) and presents the objects in a single
coordinate system. vmc uses superposition as the default
comparative layout if users don’t specify a comparative layout.
mpg_model %>%
mcplot() +
mc_layout_superposition() +
mc_gglayer(coord_flip())
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.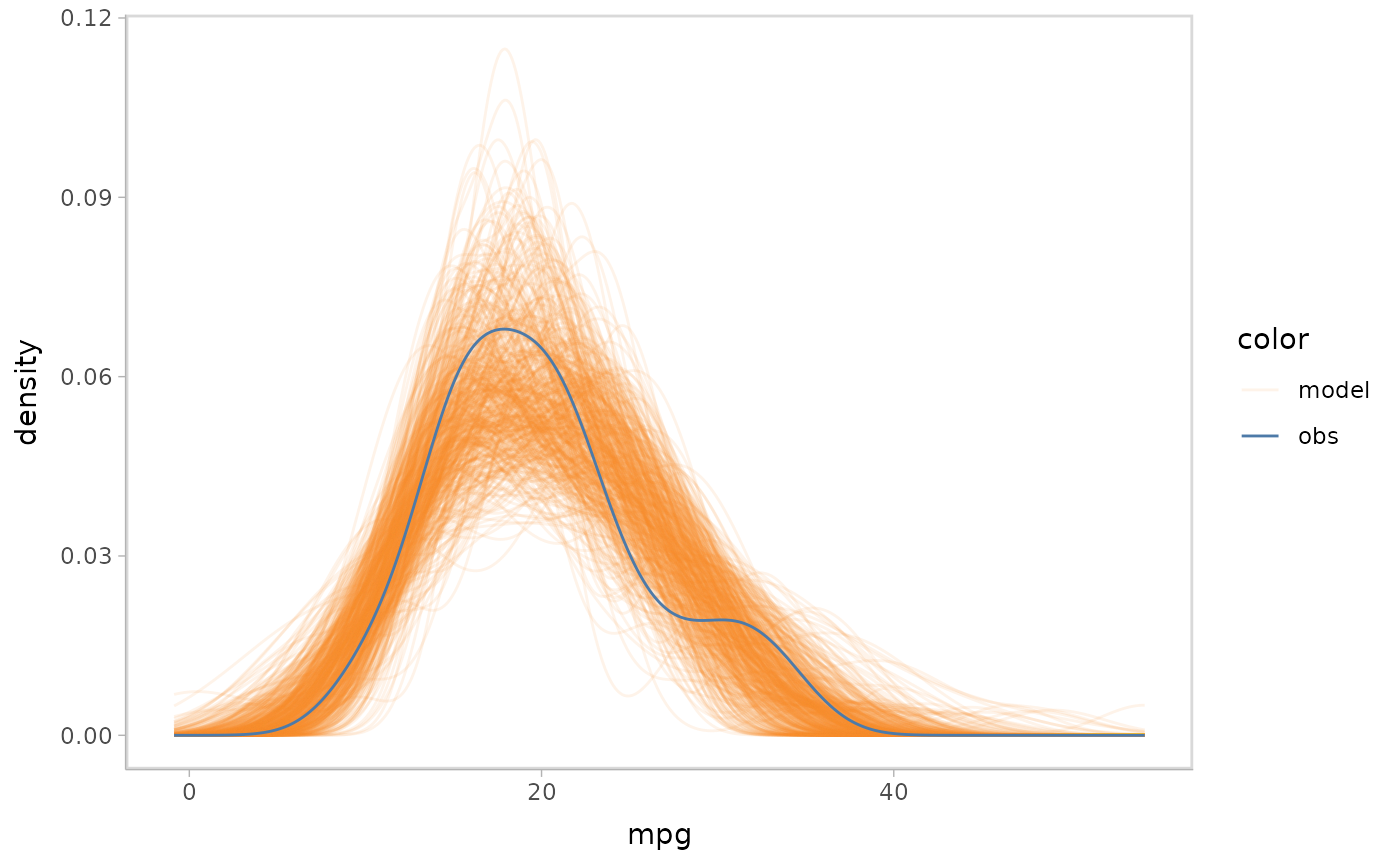
Explicit encoding
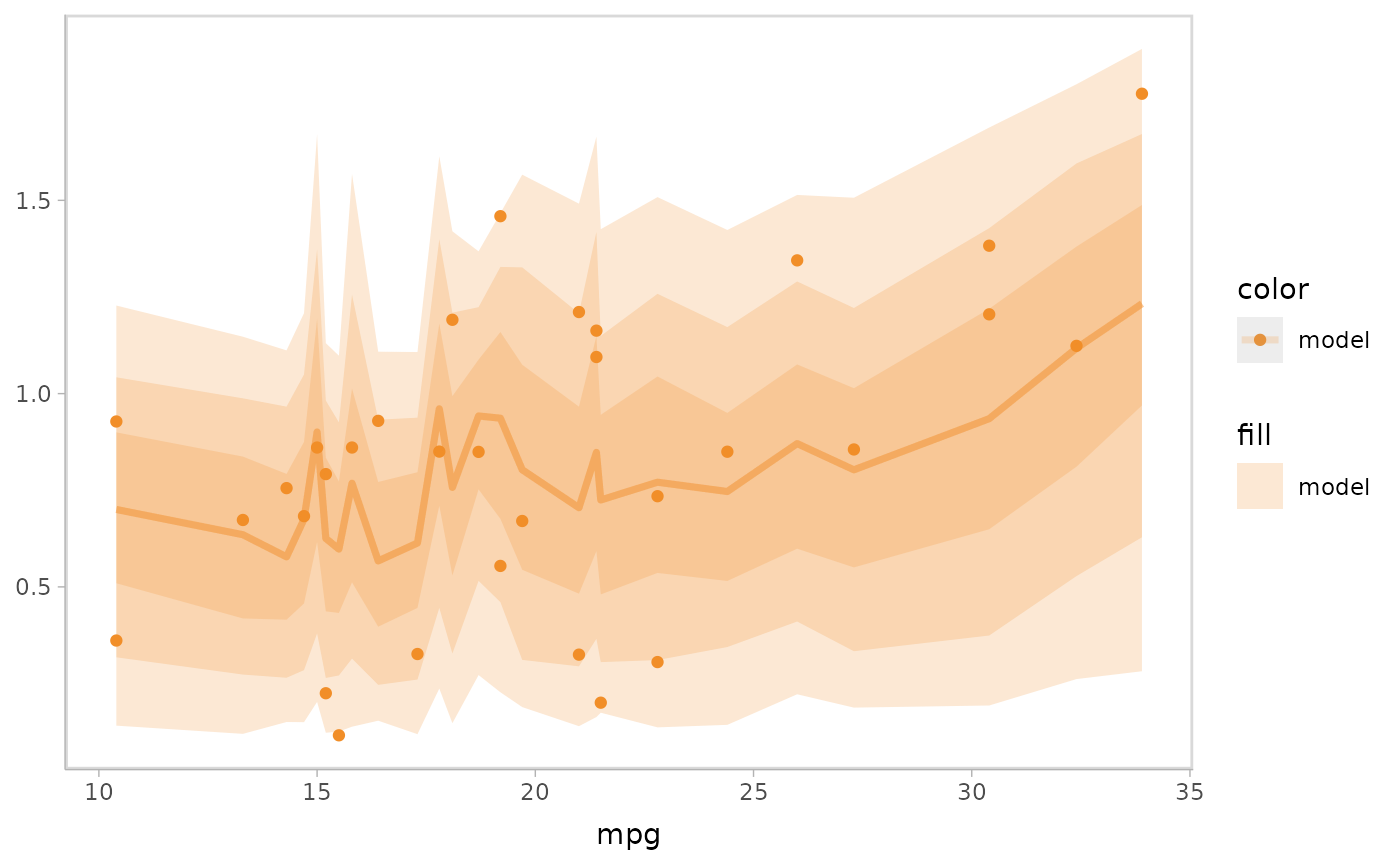
Explicit encoding directly encodes the connections between objects visually after calculating the difference between model predictions and observed data. For example, a residual plot presents the difference between the model predictions and observed data to encode the comparison about a central line encoding perfect prediction. A quantile-quantile (QQ) plot computes the quantiles of the residuals to encode the comparison.
Explicit encoding can also explicitly enable the user to check a model feature but not depends on viewer’s perception on the distribution of model prediction and observed data shown in raw coordinates of response variable and independent variables. For example, residual plot conditional on response variable enables to check linearity and QQ plot enables to check the normality of residuals.
Let’s check the linearity of the model by specifying a residual plot
conditional on the response variable mpg, where we use two
visual representations (mc_model_lineribbon() and
mc_model_point()) for the model predictions to reveal the
trend of residuals while also showing a set of raw data points.
mpg_model %>%
mcplot() +
mc_condition_on(x = vars(mpg)) +
mc_layout_encoding("residual") +
mc_model_lineribbon(alpha = .2) +
mc_model_point(n_sample = 1) +
mc_gglayer(geom_hline(yintercept = 0))
#> Warning: Duplicated aesthetics after name standardisation: alpha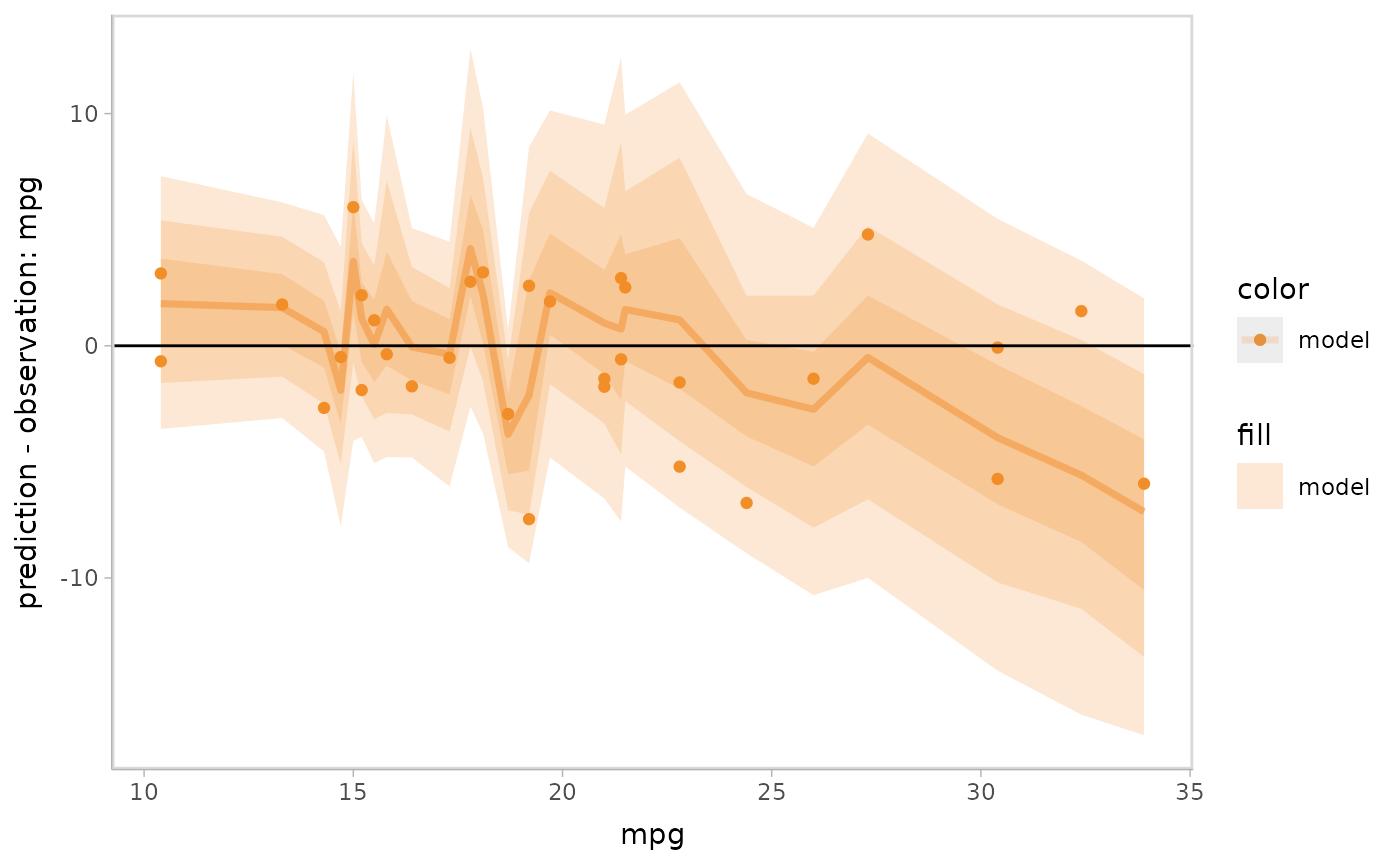
Next let’s check the normality of the residuals by drawing a QQ plot.
We could specify this easily by using
mc_layout_encoding(tranform = "qq").
mpg_model %>%
mcplot() +
mc_layout_encoding("qq") +
mc_model_point(n_sample = 1) +
mc_gglayer(geom_abline())
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.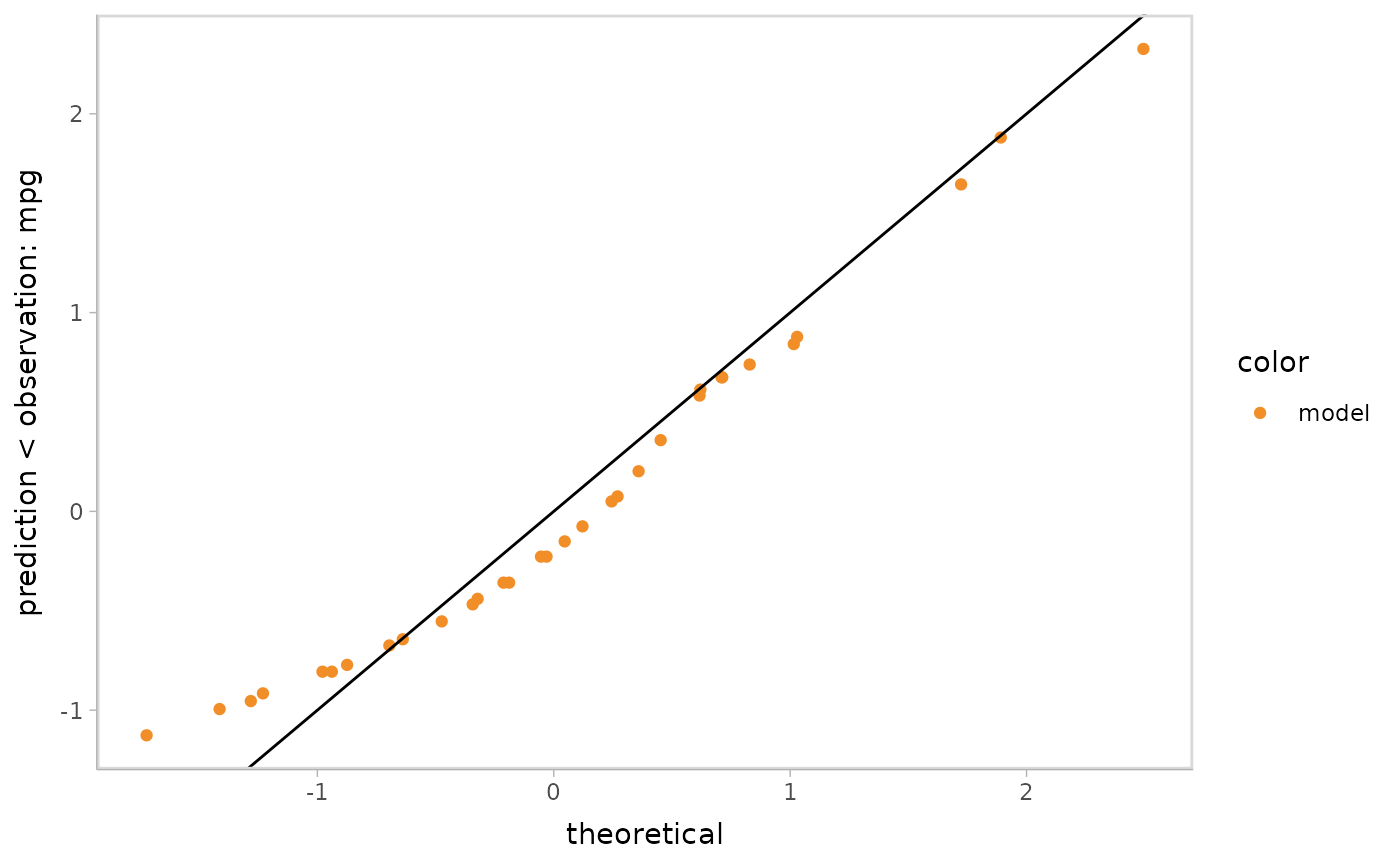
Specify your own explicit encoding
We could use vmc to specify a customized transformation
function that computes the comparison between model predictions and
observed data. The transformation function should take as input a data
frame that is generated from the newdata data frame in
mc_draw() (if not specified, it’s the data used to fit the
model, e.g., you can get that by insight::get_data(model))
with additional columns: .row, .draw,
prediction, and observation (if
newdata has a column for the response variable). We can
transform the input data frame to generate a new one that has the
columns for the variable shown on the y-axis and x-axis named as
y_axis and x_axis (optional).
Here is an example of a customized explicit encoding, where we want to check the variance of residuals.
var_res_func = function(data) {
data %>%
mutate(y_axis = prediction - observation) %>%
mutate(y_axis = sqrt(abs(y_axis / sd(y_axis))))
}
mpg_model %>%
mcplot() +
mc_condition_on(x = vars(mpg)) +
mc_layout_encoding(var_res_func) +
mc_model_lineribbon(alpha = .2) +
mc_model_point(n_sample = 1)
#> Warning: Duplicated aesthetics after name standardisation: alpha